Introduction
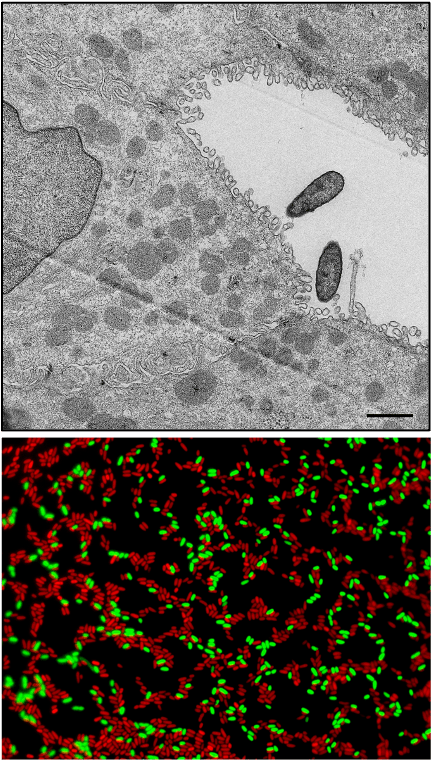
Pseudomonas aeruginosa is a widespread and highly diverse species of bacteria. It is responsible for many types of severe acute infections and is commonly isolated from the chronically infected lungs of individuals with cystic fibrosis. Of greater concern is the rapid emergence of resistance to multiple classes of antibiotics making it a serious threat to public health.
P. aeruginosa is considered a highly opportunistic bacterial pathogen. This means it takes advantage of weaknesses in our natural defense barriers to initiate an infection. When successful, it uses an arsenal of weapons (a.k.a. “virulence factors”) to protect itself from killing by our immune system. It also produces weapons to kill other bacteria! Our goal is to identify these weapons and determine how they work.
Our research spans the areas of Microbiology, Molecular Biology, Genomics, and Immunology. It is our hope that findings from this work will stimulate the development of novel treatment options against severe P. aeruginosa infections.
Contact Dependent growth Inhibition (CDI) toxins
CdiA is a large exoprotein that is known to mediate a form of bacterial competition termed Contact-Dependent growth Inhibition (CDI). CdiA forms a long filamentous stalk that facilitates delivery of its own C-terminal toxic effector domain into targeted cells (see model to the right). The enzymatic activity of the effector domain in a susceptible target bacterium results in growth arrest or cell death. CdiA-producing cells are protected from sibling intoxication by the production of a cognate immunity factor that neutralizes the effector domain. Through genomics studies we know that the CdiA effector domain and cognate immunity factor vary among different strains of P. aeruginosa. Such diversification is thought to promote kin selection and facilitate niche competition in the environment.
Interestingly, we have discovered that certain CdiA-Tox domains mediate both interbacterial competition and virulence in P. aeruginosa. Our research uses bacterial genetics, biochemistry, cell biology, and infection models to understand how a system used in bacterial competition may serve a dual role in the pathogenesis of P. aeruginosa
FIGURE: CdiA Model (of bacterial competition) – CdiA forms a long stalk that presents a receptor binding domain (RBD) distal to the cell surface of the attacking bacterium. Once the RBD interacts with a susceptible target bacterium, the remaining carboxy-domains facilitate translocation of a toxic C-terminal effector domain into the target bacterium.

Rearrangement Hot Spot (RHS) toxins
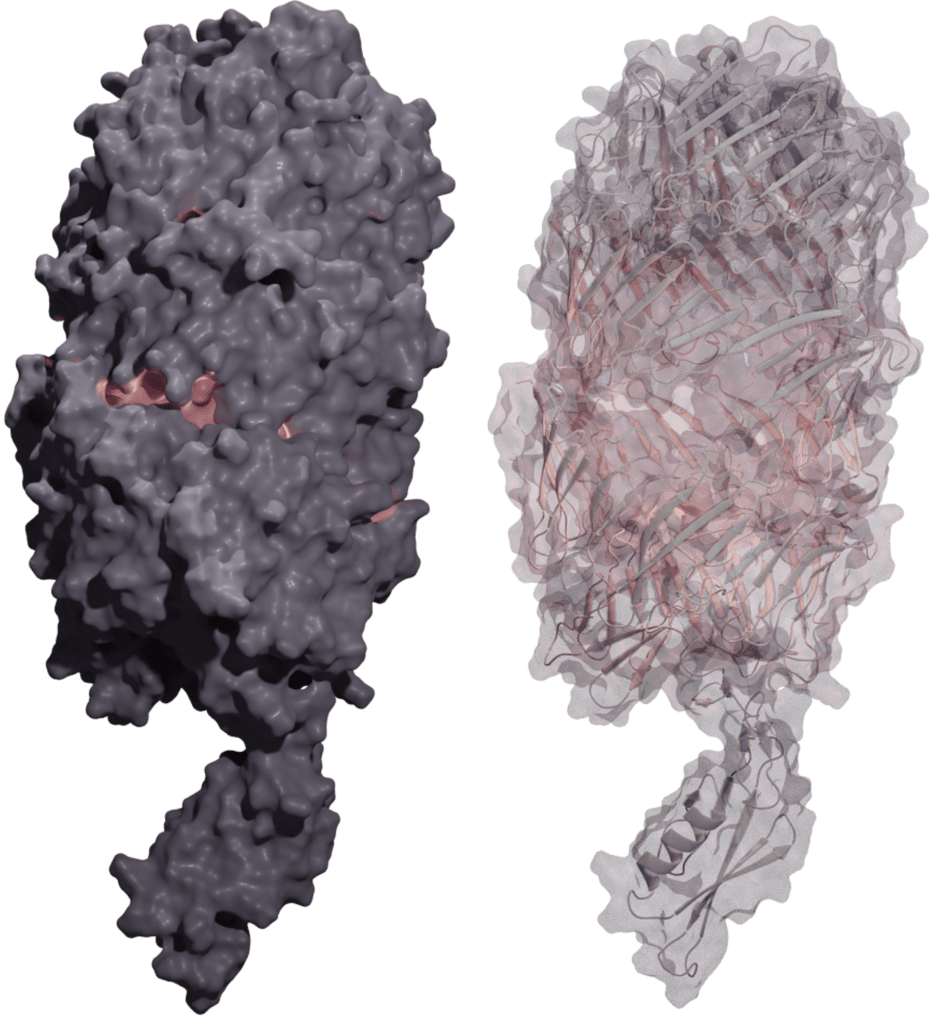
We are studying the role of a P. aeruginosa protein pair that we refer to as PxrB and PxrA (Polymorphic toXin with Rhs elements). These two proteins contain RHS elements, a series of tyrosine-aspartate (YD) repeating motifs, that are predicted to associate together and form a shell around a small toxic effector domain (left). There are some general similarities of the PxrBA toxin to CdiA: (1) The enzymatic activity of the effector domain in a susceptible target bacterium results in growth arrest or cell death; (2) Toxin-producing cells are protected from sibling intoxication by the production of a cognate immunity factor that neutralizes the effector domain; (3) Through genomics studies we know that the PxrA effector domain and cognate immunity factor vary among different strains of P. aeruginosa; and (4) We have discovered that PxrBA both mediates interbacterial competition and influences virulence. We are currently investigating the PxrBA toxin for its mechanism of action, toxic effect on cells, and role in P. aeruginosa virulence. We are also interested in engineering this system to deliver potent antimicrobials as an alternative to antibiotic therapies.
FIGURE: PxrBA toxin Model – AlphaFold prediction of the PxrBA holotoxin. The toxic effector domain is contained within the interior of the shell. Image created with blender.
Infection Dynamics
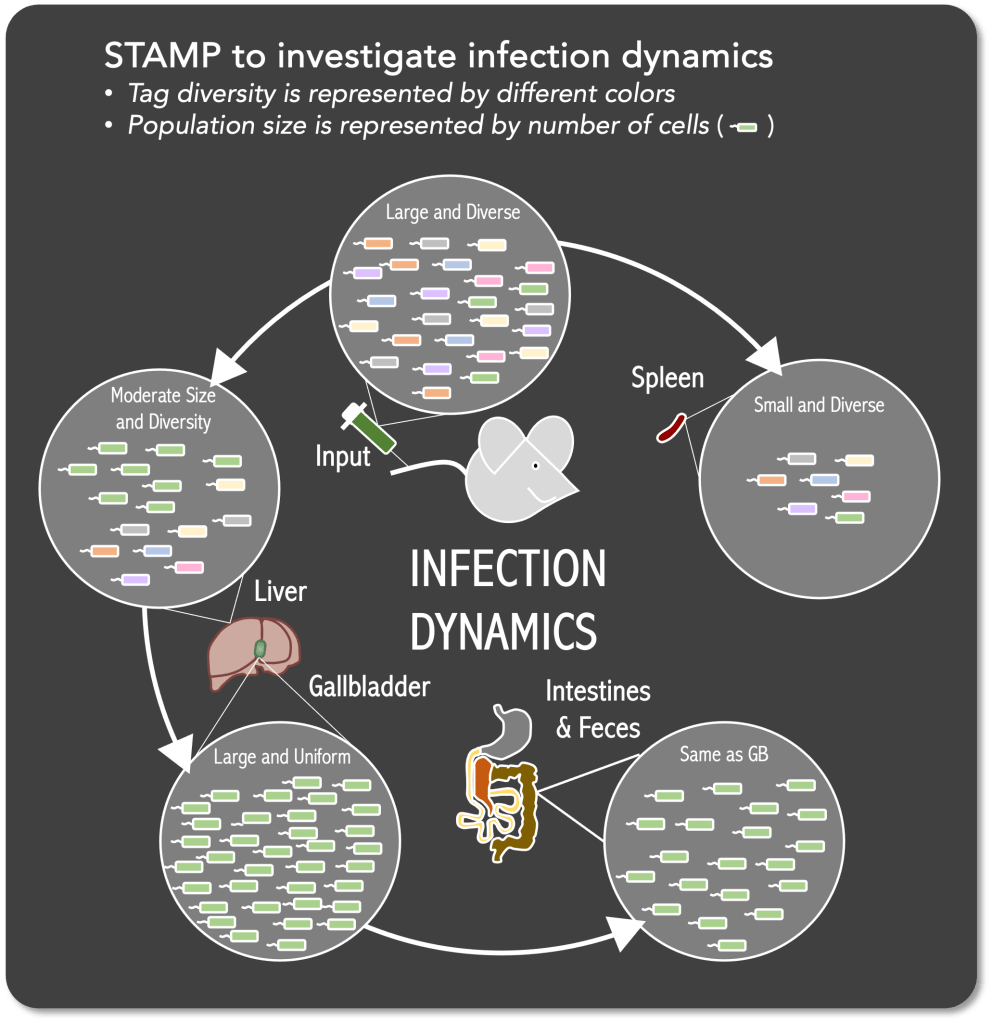
“Infection Dynamics” during invasive disease refers to understanding (1) where the bacteria go, (2) how they get there, (3) what bacterial factors are involved, and (4) any host bottlenecks the bacteria must overcome.
With Dr. Kelly Bachta, https://sites.northwestern.edu/bachtalab/, we have a longstanding collaboration characterizing the dynamics of invasive P. aeruginosa infections. Using a variety of imaging and genomics-based tools, we have determined that an initial population of disseminating bacteria escape clearance in the spleen and liver. A small subpopulation of bacteria makes their way to the gallbladder, likely from the liver through bile ducts. Once in the gallbladder, robust bacterial replication occurs. This massive replication promotes bacterial dissemination to the intestinal tract and excretion in the feces. This type of bacterial escape has substantial clinical implications concerning transmission and secondary infection.
We will continue to investigate the consequence of bacterial dissemination to the billiary and intestinal tracts, and elucidate host-microbe interactions at the spleen and liver. We anticipate this work to highlight key breakpoints during P. aeruginosa infection that can be targeted for specific therapies.
FIGURE: Cartoon diagram of a genomic barcoding approach to quantify population bottleneck effects in vivo. This process termed STAMP (sequence tag-based analysis of microbial population dynamics) tracks changes to an infecting population’s richness and evenness during the course of an infection.